Abstract
Introduction: Acute myeloid leukemia (AML) is a disease characterized by the expansion of myeloid blasts in the bone marrow. Recently, we published the results of the first proteogenomic characterization of bone marrow blasts from 177 adult patients with AML (10.1016/j.ccell.2022.02.006) that were uniformly treated with intensive induction chemotherapy. Using mass spectrometry (MS) and next-generation sequencing we curated a multi-omics data set with global protein and gene expression, DNA mutations and chromosomal aberrations. We identified a proteomic subtype characterized by enhanced expression of mitochondrial proteins, significantly poorer overall survival and high sensitivity to inhibitors targeting mitochondrial respiration (Mito-AML). We have now expanded the proteomic data set with bone marrow biopsies from 90 additional adult patients with AML in order to facilitate the proteomic characterization of individual genetic subtypes of AML including nucleophosmin 1 (NPM1)-mutated AML.
Methods: We used a label-free (LF), data-independent (DIA) MS-based proteomic workflow to profile protein expression in bone marrow-derived leukemic blasts from 90 additional patients with AML. The patient cohort was similar to our initial cohort with regard to demographic-, disease- and clinical characteristics. We applied stringent filtering, missing value imputation and batch correction to merge the two cohorts. We were able to validate the proteomic Mito-AML subtype in this merged cohort by our previously published machine-learning model. Next, we investigated proteomic differences between NPM1mut (n=89) and NPM1wt (n= 172) patients using linear models; we identified differentially regulated biological processes from the gene ontology (GO) and REACTOME gene sets as well as gene expression signatures of healthy and malignant cellular subtypes (van Galen et al.) using a gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA). Clustering of the NPM1mut proteome was done using a consensus approach on the Euclidean distances and kmeans with k = 2 was chosen based on a set of metrices, including silhouette and concordance score.
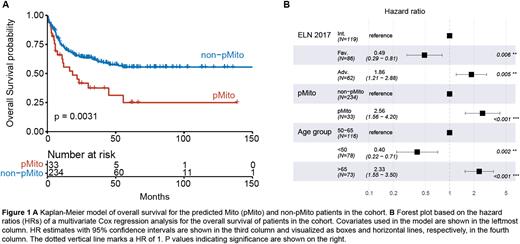
Results: The global protein expression structure, represented by inter-sample Euclidean distance, was largely independent of blast maturity, genetic risk groups and mutations. In our combined cohort of 267 patients, we identified 33 Mito-AML patients, confirming a frequency of 12 % of this proteomic AML subtype. As published by us previously, Mito-AML showed poor overall survival and independent, significant risk of death in the combined cohort as well as in the 90 patient sub-cohort.
Mutations in NPM1 are frequently found in de novo AML and are characterized by a more chemo-sensitive disease. In addition to proteins previously described as being differentially expressed we identified a number of previously unreported proteins including surface proteins (e.g. DmX-like protein 2 (DMXL2)), to be significantly higher expressed in NPM1mut AML. Together with our proteomic data for CD34+ progenitor cells from healthy donors our resource data set might reveal potential target structures for cellular therapies. Among the gene sets differentially enriched between NPM1mut and NPM1wt were terms associated to mitochondrial respiration and metabolism. Interestingly, Mito-AML was enriched among NPM1mut AML (48.5% in NPM1mut vs. 32% in NPM1wt), indicating that Mito-AML is a high-risk subset within NPM1mut AML. Furthermore, we identified regulation of chromosomal and nucleosome architecture amongst the top down-regulated processes in NPM1mut which is in accordance with the role of wild-type NPM1 to chaperone these structural processes. Interestingly, we observed among NPM1mut AML two sub-clusters that were either enriched for blasts with myelo-monocytic differentiation or for progenitor- and stem cell-like blasts indicating a cell-of-origin phenotype.
Conclusion: We report the expansion of our previously published proteomic characterization of AML with bone marrow-derived AML blasts from 90 additional patients. We confirm the proteomic Mito-AML subtype and its poor outcome under intensive chemotherapy. We furthermore elucidate the proteomic landscape of NPM1mut AML and identifie two subgroups of NPM1mut AML that are characterized by either myelo-monocytic differentiation or a progenitor and stem cell-like blast state.
Disclosures
Thiede:AgenDix GmbH: Current Employment, Current equity holder in private company; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Janssen Pharmaceuticals: Speakers Bureau; Jazz Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Kronos Bio, Inc.: Honoraria. Steffen:AbbVie: Other: Travel/Congress Participation Support; Jazz Pharmaceuticals: Other: Travel/Congress Participation Support. Stegmaier:Novartis: Research Funding; KronosBio: Consultancy, Research Funding; AstraZeneca: Consultancy; Auron Therapeutics: Consultancy, Current holder of stock options in a privately-held company, Membership on an entity's Board of Directors or advisory committees. Oellerich:Merck KGaA: Consultancy, Research Funding; Gilead Sciences, Inc.: Research Funding; F. Hoffmann-La Roche Ltd: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal